How do I do that?

Best answer by Allie
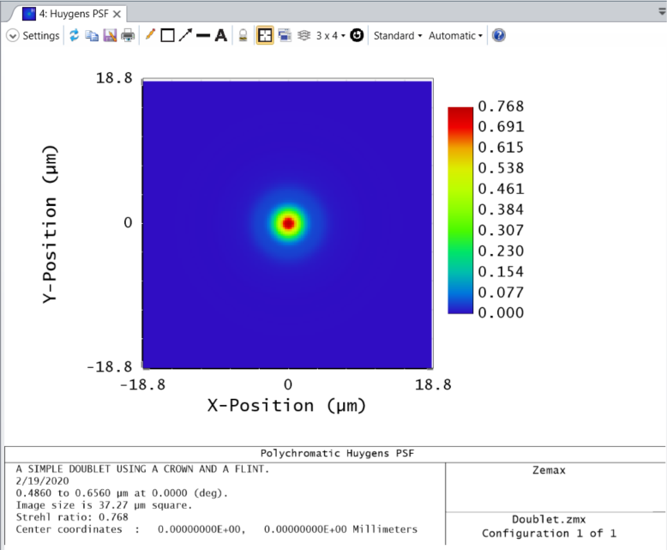
These types of plots (Huygen's PSF, Wavefront Map, Detector Viewer, etc.) are generated with a matrix of data points provided within the "Text" tab of the same analysis window. This matrix data may be pulled into the API using the ZOSAPI.Analysis.Data.IAR_ Interface and made into a figure.
Analysis window data is split into multiple categories within the IAR_ Interface. A matrix of data (shown above) will typically be provided as a DataGrid while a 2 column list of data (such as that given by a Cross Section plot) will be provided by a DataSeries. Sometimes, analysis window results are not pulled into the API, in which case we must pull the results into an external text file. See "Generating a list of output data types for each analysis in the ZOS-API" for more information.
As long as the data is accessible, we can pull it into the API using the corresponding IAR_ Interface property. Below is a Matlab example of this process:
TheSystem = TheApplication.PrimarySystem;
% Open the Huygen's PSF. We will use the default settings for now
huygensPSF = TheSystem.Analyses.New_HuygensPsf();
% Run the analysis with the current settings and pull the results
huygensPSF.ApplyAndWaitForCompletion();
huygensResults = huygensPSF.GetResults();
% The results will be split into multiple data structures
% One structure will house the header information
% Another structure will house the relative intensity values
% Pull the structure with the intensity values
matrixData = huygensResults.DataGrids(1).Values.double;
% OpticStudio has pixel (1,1) in the top left of the matrix
% Matlab places the (1,1) at the bottom so we need to flip the matrix
huygensData = flipud(matrixData);
% Use pixel data to create a figure
% The jet colormap will match closely with the False Color plot
imagesc(huygensData)
colormap jet;
axis square;
% Close the Huygen's plot
huygensPSF.Close();
Enter your E-mail address. We'll send you an e-mail with instructions to reset your password.